Indice del volumen
Volume index
Comité Editorial
Editorial Board
Comité Científico
Scientific Committee
- Step 1 (3D representation of compounds)
The three-dimensional representations of twenty 1,3,4-thiadiazole disulfonamide and twenty 1,3,4-thiadiazoline disulfonamide compounds were building up by the use of HyperChem software32. - Step 2 (creation of measured properties file)
The measured inhibition concentration of each compound transformed to logarithmic scale was stored in property.txt file. - Step 3 (generation of molecular descriptors family)
All forty compounds were used in construction and generation of the molecular descriptors family. The algorithm of molecular descriptors family list generation for studied substituted thiadiazole- and thiadiazoline-disulfonamide was strictly based on compounds structure. In order to discard redundant information a bias algorithm with a significance level of 10-9 was applied after molecular descriptors family generation. Each descriptor from family has an individual seven-letters name which expresses the modality of construction. The 7th letter indicates the compound characteristic relative to its geometry (g) or topology (t); the 6th letter, the atomic property (cardinality - C, number of directly bonded hydrogen’s - H, atomic relative mass - M, atomic electronegativity - E, group electronegativity - G, partial charges - Q); the 5th letter indicates the atomic interaction descriptor; the 4th letter indicates the overlapping interaction model; the 3rd letter denotes the fragmentation criterion (the minimal fragments - m, the maximal fragments - M, the Szeged fragments criterion - D, and the Cluj fragments criterion - P 33,34); the 2nd letter refer the cumulative method of fragmentation properties (one out of nine selectors 29), and the 1st letter indicates the linearization procedure applied in generation of global molecular descriptor (identity - I, inverse - i, absolute - A, a inverse of absolute - a, natural logarithm of absolute value - L, simple natural logarithm - l). - Step 4 (search and identification of MDF- SAR models)
The criterions imposed in search and identification of the MDF-SAR models were the highest values for the correlation coefficient and for the squared correlation coefficient (close to +1 or -1). - Step 5 (MDF-SAR models validation)
Analysis of predictive abilities of the chosen MDF-SAR multi-varied models was performed by computing the following parameters: cross-validation leave-one-out (loo) scores (r2cv-loo), Fisher parameter (Fpred) and associated significance (ppred), and standard error (sloo). In leave-one-out analysis the property of each compound was predicted by the regression equation calculated based on the all other compounds. The internal predictive analysis of MDF-SAR models was performed with Leave-one-out Analysis application35. - Step 6 (MDF-SAR models analysis)
The chosen MDF-SAR models were analyzed through computing and interpreting the following parameters: the correlation coefficient (r), the squared correlation coefficient (r2), the adjusted squared correlation coefficient (r2adj), the standard error of estimation (sest), the Fisher parameter (Fest) and its significance (pest), the 95% confidence intervals (95%CIc) of multi-varied regression coefficients, the Student parameter (tdescriptor) and associated significance, the parameters of co-linearity (analysis of the squared correlation coefficients between descriptors, r2(descriptor, descriptor), and between each descriptor and measured IC50 activity r2(log IC50, descriptor)), and the model stability (defines as the difference between squared correlation coefficient and leave-one-out correlation coefficient score (r2 - r2cv-loo); the model was consider stable if the difference had a lower value).
MODELLING THE INHIBITORY ACTIVITY ON CARBONIC ANHYDRASE IV OF SUBSTITUTED THIADIAZOLE - AND THIADIAZOLINE - DISULFONAMIDES: INTEGRATION OF STRUCTURE INFORMATION
Lorentz Jäntschi and Sorana Daniela Bolboaca*
Technical University of Cluj-Napoca
*"Iuliu Hatieganu" University of Medicine and Pharmacy
Cluj-Napoca, Romania
lori@academicdirect.org
Rev Electron Biomed / Electron J Biomed 2006;2:22-33
Comment of the reviewer Jose Luis Hernandez Caceres, PhD. Full Professor. Center for Cybernetics Applications to Medicine (CECAM). Havana, Cuba.
Comment of the reviewer Prof.Dr. Amalio Garrido Escudero Chem Eng PhD. Head Environmental Engineering and Toxicology Dpt. Universidad Católica S. Antonio. Campus Los Jerónimos. Guadalupe (Murcia). SPAIN
ABSTRACT:
Purpose: To analyze the relationships between inhibitory activities on carbonic anhydrase IV and structures of substituted 1,3,4-thiadiazole and 1,3,4-thiadiazoline disulfonamide through integration of compounds complex structure information by the use of Molecular Descriptors Family.
Method: A number of forty compounds were used to generate and compute the molecular descriptors family and to build structure-activity relationships models. The obtained multi-varied models (the models with two, respectively with four descriptors) were validated by computing the cross-validation leave-one-out score (r2cv-loo), and analyzed through assessment of the squared correlation coefficients (r2), and the models stability (r2 - r2cv-loo). The estimation abilities of the multi-varied MDF-SAR model with four descriptors were analyzed in training and test sets.
Results: Analysis of the obtained models shows that the best results was obtained by the multi-varied model with four molecular descriptors (r2 = 0.920). The prediction abilities of this model is sustained by the cross validation leave-one-out score (r2cv-loo = 0.903), the model stability (r2 - r2cv-loo = 0.017), and the results on training versus test analysis (no significant differences between correlation coefficients in training and test sets, p > 0.05). The multi-varied model which used four descriptors proved to render higher value of correlation coefficient comparing with previous reported models (p < 0.01).
Conclusion: The multi-varied model with four descriptors is a solid and reliable one and indicates that the inhibition activity on carbonic anhydrase IV of substituted 1,3,4-thiadiazole- and 1,3,4-thiadiazoline-disulfonamides is like to be of geometry and topology nature, being in relation with compounds partial charges.
KEYWORDS:Molecular Descriptors Family (MDF); Carbonic anhydrase IV inhibitors; Substituted 1,3,4-thiadiazole- and 1,3,4-thiadiazoline-disulfonamides
RESUMEN:
MODELIZACIÓN DE LA ACTIVIDAD INHIBIDORA DE LA CARBOANHIDRASA IV PRODUCIDA POR LAS DISULFONAMIDAS SUSTITUIDAS DEL TIADIAZOL Y DE LA TIADIAZOLINA: INTEGRACIÓN DE LA INFORMACIÓN
ESTRUCTURAL.
Objetivo: Objetivo: Analizar las relaciones entre las actividades inhibidoras en la carboanhidrasa IV y las estructuras de las sulfonamidas sustituidas del 1,3,4-tiadiazol y de las disulfonamidas sustituidas de la 1,3,4-tiadiazolina mediante la integración compleja de la información estructural de los compuestos por el uso de Familias de Descriptores Moleculares.
Método: Un número de cuarenta compuestos fue utilizado para generar y para calcular las familias de descriptores moleculares y para construir modelos de las relaciones estructura-actividad. Los modelos multivariantes obtenidos (con dos y cuatro familias de descriptores respectivamente) fueron validados calculando la validación cruzada
"leave-one-out" (r2cv-loo), y analizadola mediante la evaluación de los coeficientes de correlación ajustados (r2), y la estabilidad de los modelos (r2 - r2cv-loo). Las capacidades de estimación del modelo multivariante de MDF-SAR con cuatro descriptores fueron comprobadas en los conjuntos de datos experimentales y previstos por el modelo.
Resultados: El análisis de los modelos obtenidos demuestra que los mejores resultados fueron obtenidos por el modelo multivariante con cuatro descriptores moleculares (r2 = 0.920). La capacidad de predicción de este modelo son sostenidas por la validación cruzada "leave-one-out" r2cv-loo = 0.903, la estabilidad del modelo (r2 - r2cv-loo = 0.017), y los resultados entre valores predichos por el modelo frente a valores experimentales (sin diferencias significativas entre los coeficientes de correlación en los conjuntos de datos, p > 0.05). El modelo multivariante que utilizó cuatro descriptores mostró un valor más alto del coeficiente de correlación en comparación con los modelos divulgados anteriormente (p < 0.01).
Conclusión: El modelo multivariante con cuatro descriptores es sólido y fiable e indica que la actividad de la inhibición en la carboanhidrasa IV producida por las sufonamidas sustituidas del 1,3,4-tiadiazol- y de la 1,3,4-tiadiazolina- dependen de la naturaleza de la geometría y de la topología del compuesto, estando relacionadas con las cargas parciales de los mismos.
PALABRAS CLAVES: Familia de Descriptores Moleculares (MDF); inhibidores de la carboanidrasa IV; Sulfonamidas sustituidas del 1,3,4-tiadiazol- y la 1,3,4-tiadiazolina.
INTRODUCTION
Quantitative structure-activity relationships (QSAR) is widely use in modelling of physico-chemical properties or biological activities of chemically active compounds1-3, in most cases by the used of topological indices as: Szeged index4,5, Wiener index6-8, PI (Padmakar - Ivan) index9,10, Balaban index11, Zagreb index12, valence connectivity index13, Lu index14, DAI index (distance-based atom-type topological)15, AI index (atom-type topological index)16. Carbonic anhydrase IV, a membrane-bound form of carbonic anhydrase enzymes, is known to be, comparing with carbonic anhydrase II, a faster intra-molecular proton transporter17. Carbonic anhydrase inhibitors were studied by many authors through quantitative structure-activity relationships18-24. More, the studies of carbonic anhydrase IV inhibition with sulfonamide lead to development of drugs used as antiglaucoma agents25, anticonvulsant26 agents, and as novel types of anticancer agents27.
A number of forty substituted disulfonamide, twenty 1,3,4-thiadiazole disulfonamide and twenty 1,3,4-thiadiazoline disulfonamide, with carbonic anhydrase IV inhibition properties were previously studied by the use of quantum chemical quantitative structure-activity relationships28. The models obtained by Supuran&Clare are in Table 1, and associated statistics in Table 2. In table 1, there were used the following abbreviations: IIzz = the polarizability tensor, QCr1 = the changes of the atoms of the attached ring carbon, QS1 = the changes of the atoms of the primary sulfonamide group, µx = the dipole moment, QNr1 = the charges on the nitrogen and ΔHS2 = the solvation energy for the secondary sulfonamide group28.

Table 1. QSAR models for carbonic anhydrase IV inhibition activity
In the table 2 were used the following abbreviations: n for the studied sample size, R2 for the square of the multiple correlation coefficient, Q2 for the same quantity base on the predicted errors (the leave-one-out techniques), s for the standard errors of estimate of the equation, and F for the Fisher variance ratio.

Table 2. Statistics of models recorded in table 1
Starting from the successful results obtained by an original molecular descriptors family (MDF) on structure-activity relationships (SAR) methodology29-31, the aim of the research was to investigate and to assess the estimation and prediction abilities of the MDF-SAR methodology on a sample of substituted thiadiazole- and thiadiazoline-disulfonamides.
MATERIALS AND METHODS
Carbonic anhydrase IV inhibitors
A sample of twenty 1,3,4-thiadiazole disulfonamides and twenty 1,3,4-thiadiazoline disulfonamides, with carbonic anhydrase IV inhibition properties was included into the study. The measured inhibitory activity of compounds, expressed as logarithm of concentration of the agent that is required for fifty percent inhibition in vitro (log IC50), was taken from a previous study28.
The generic structure of compounds, the compounds abbreviation, substituents and compounds measured inhibition activity are in Table 3.
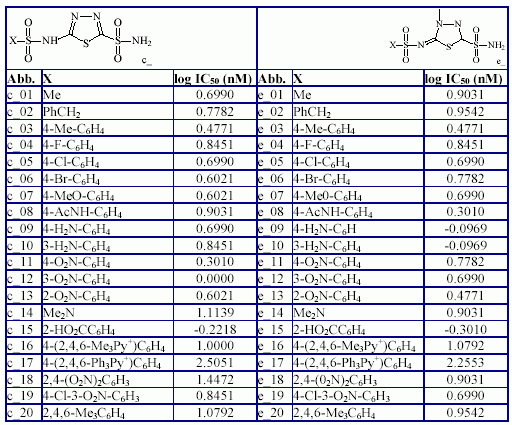
Table 3. Structure of compounds and measured IC50
Modelling the inhibition activity of substituted thiadiazole- and thiadiazoline disulfonamide
The inhibition activity of substituted thiadiazole- and thiadiazoline- disulfonamide were modelled through integration of complex information obtained from compounds structure. The applied methodology used an original molecular descriptors family approach in order to estimate and predict the inhibition activity of studied substituted thiadiazole- and thiadiazoline-disulfonamide.
The steps applied in MDF-SAR modelling29 were:
The comparison between multi-varied model with two and respectively four descriptors was performed through a correlated correlation analysis by the use of Steiger test36.
The comparison between the correlation coefficients of previous reported models and the correlation coefficients obtained by MDF-SAR multi-varied models was analyzed by the use of Fisher's Z test. The estimation abilities of the model with the highest squared correlation coefficient was analyzed in training and test sets with Training vs. Test application37. There were analyzed twelve situations, starting with sample sizes in training set from twenty to thirty-one and corresponding sample sizes in test set from twenty to nine. For each training and test set, the application rebuilds the regression model and based on the obtained model the activity of compound from test set was predicted.
RESULTS
Two MDF-SAR multi-varied models, one with two and other with four molecular descriptors, proved to have abilities in estimation and prediction of 1,3,4-thiadiazole disulfonamide and 1,3,4-thiadiazoline disulfonamides inhibitory property. The equations of the MDF-SAR models are:
Ŷ2d = 0.802+0.111·inPRlQg+9.980·10-9·iHMMTQt (1)
Ŷ4d = 0.625+0.105·inPRlQg+9.919·10-9·iHMMTQt-9.248·IHMDTQg+1.727·InPdJQg (2)
The molecular descriptors used by the models, their calculated values and the values of the inhibition concentration 50% (IC50) estimated by each models (Ŷ2d respectively Ŷ4d) are in Table 4.
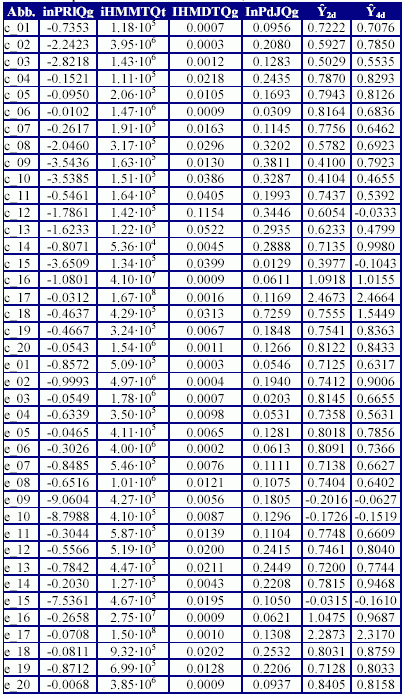
Table 4. Descriptors used in MDF-SAR models, their values and estimated IC50
The statistics of the MDF-SAR models are in Table 5 and Table 6.
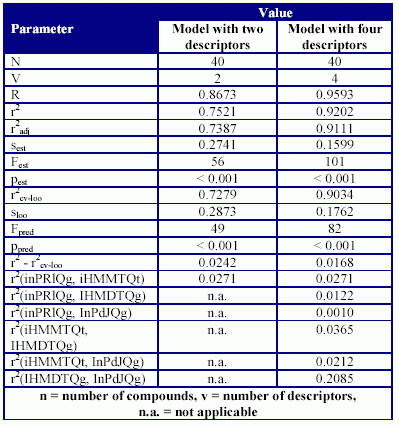
Table 5. The statistical parameter of MDF-SAR models
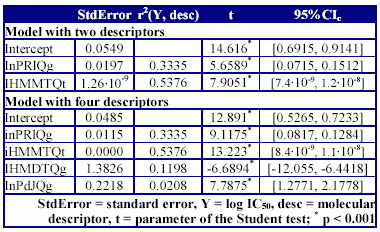
Table 6. The qualities of MDF-SAR models
The plots of residuals (defined as the difference between the measured IC50 and estimated by the regression equations (1) and (2)) obtained with the MDF-SAR models with two and respectively four molecular descriptors are in Figure 1.
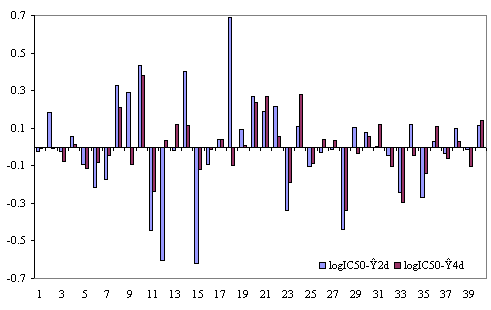
Figure 1. The plots of residuals obtained with Eq. (1) and (2)
The graphical representation of the measured IC50 versus estimated with MDF-SAR model with four descriptors (Eq.(2)) is in Figure 2.
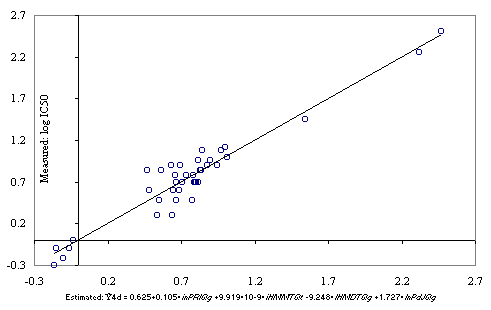
Figure 2. Estimated with the Eq. (2) versus measured IC50 of substituted thiadiazole- and thiadiazoline-disulfonamide
A correlated correlation analysis was used in order to test the hypothesis that the correlation coefficient obtained by the MDF-SAR model with four descriptors is not statistical different comparing with the correlation coefficient obtained with the MDF-SAR model with two descriptors. The results expressed as correlation coefficients, parameter of the Steiger's Z test and associated significance are in Table 7.
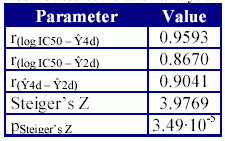
Table 7. The results of the correlated correlation analysis
Comparing the performances of the MDF-SAR model with two descriptors (Eq. (1)), respectively with four descriptors (Eq. (2)) with previous reported models (model 1 and 2, table 2), there was observed that the MDF-SAR model with four variables obtained a correlation coefficient statistically grater (Eq.(2)-model 1, pSteiger’s Z = 1.38∙10-4; Eq.(2)-model 2, pSteiger’s Z = 8.40∙10-3).
The results obtained in training versus test analysis, applied for the MDF-SAR model with four descriptors are in Table 8, and the plot of squared correlation coefficients in Figure 3. The abbreviations used in Table 8 had the following significances: the number of training versus test analysis (No.), the coefficients (a0, a1, a2, a3, and a4) of the generic MDF-SAR model with four descriptors (Ŷ4d = a0 + a1·inPRlQg + a2·iHMMTQt + a3·IHMDTQg + a4·InPdJQg), the number of compounds in training (Notr) respectively in test (Nots) sets, the correlation coefficients obtained in training (rtr) and test (rts) sets and associated 95% confidence intervals (95%CIrtr - for training sets, respectively 95% CIrts – for test sets); the Fisher Z test for training (Ftr) respectively for test (Fts) sets, and the Fisher Z- test (FZ-test) which test the null hypothesis that there were not significant differences between correlation coefficient obtained in training set and the correlation coefficient obtained in the associated test set.
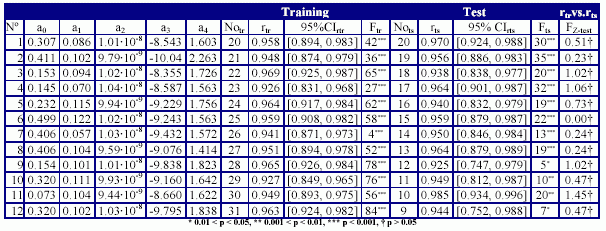
Table 8. The quality of MDF-SAR multi-varied model with four descriptors in training versus test analysis
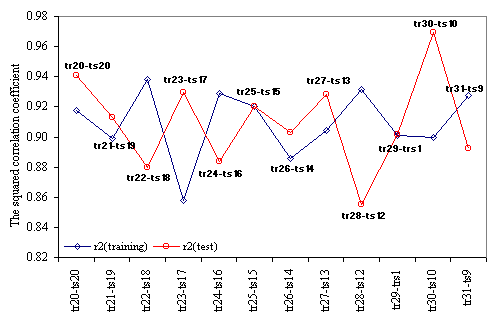
Figure 3. Training and test squared correlation coefficients
obtained with MDF-SAR model with 4 descriptors
DISCUSSIONS
The inhibition activity on carbonic anhydrase IV of substituted 1,3,4-thiadiazole and 1,3,4-thiadiazoline disulfonamides proved to be in relationship with the complex information obtained from compounds structure. In both multi-varied MDF-SAR equations (Eq.(1) and Eq.(2)) the inhibition activity on carbonic anhydrase IV was in relationship with the geometry (inPRlQg, IHMDTQg, InPdJQg) as well as the topology (iHMMTQt) of the compounds and was strongly depend on the compounds partial charges (inPRlQg, iHMMTQt, IHMDTQg, InPdJQg)
Two out of two (in MDF-SAR model with two descriptors, Eq.(1)), respectively three out of four (in MDF-SAR model with four descriptors, Eq.(2)) molecular descriptors had positive regression coefficients. Just one out of four descriptors (in MDF-SAR model with four descriptors, Eq.(2)) had a negative regression coefficient.
Analyzing the performances of MDF-SAR models it can be observed that both are significant in estimation as well as in prediction (see statistical parameters from table 5). Almost seventy-five percent in inhibition activity on carbonic anhydrase IV of studied disulfonamides can be explained by its linear relationship with the variation of two molecular descriptors (Eq.(1)). The MDF-SAR model with two descriptors proved to be valid and stable (p < 0.001; r2cv-loo = 0.7279; r2 - r2cv-loo = 0.0242).
Both descriptors used in Eq.(1) could be found again in MDF-SAR model with four variables, demonstrating that these descriptors had an important contribution in modelling of the inhibition activity on carbonic anhydrase IV of substituted 1,3,4-thiadiazole disulfonamide, 1,3,4-thiadiazoline disulfonamides. The abilities in estimation for the MDF-SAR model with four descriptors are sustained by the value of correlation coefficient (r2 = 0.9593, table 5), confidence boundaries associated with the regression coefficients and probability associated with Student tests (for all coefficients less than 0.001, see table 6). Almost ninety-six percent from variation of inhibition activity on carbonic anhydrase IV can be explained by its linear relationship with the variation of four molecular descriptors used in Eq. (2). The Fisher parameter computed in leave-one-out analysis and its significance (Fpred = 82, p < 0.001) sustains the estimation ability of the model. The stability of the MDF-SAR model (Eq.(2)) is sustained by the values of difference between correlation coefficient (r2 - r2cv(loo) = 0.0168) and by the cross validation leave-one-out correlation score (r2cv-loo = 0.9034). The power of the model with four descriptors in prediction of the inhibition on carbonic anhydrase IV of studied disulfonamides is sustained by the absence of co-linearity between descriptors (see the squared correlation coefficients between pairs of descriptors, which always is less than 0.20, table 5).
The MDF-SAR model with four descriptors proved to had a significantly greater correlation coefficient comparing with the MDF-SAR model with two descriptors (p < 0.001, table 7) and statistically significant greater correlation coefficients comparing with previous reported models (p < 0.01). The residuals of the MDF-SAR model with two descriptors varied from -0.6196 to 0.6917 while the residual of the MDF-SAR model with four descriptors varied from -0.3392 to 0.3796. The qualities of the MDF-SAR model with four descriptors in training versus test analysis showed that excepting the intercept, in more than 83%, the values of equation coefficients (a1, a2, a3, and a4) are included into the 95% confidence intervals of the model (see table 6 and table 8). More, with a single exception (for training sample size equal with thirty, table 8) the values of the correlation coefficients obtained in test sets are included into the 95% confidence intervals of the correlation coefficients obtained in training sets. There were not identify statistical significances between the correlation coefficients in training and test sets (p-value > 0.05, table 8).
Starting from the above described MDF-SAR model with four descriptors, and by the use of an original software38, the inhibition activity on carbonic anhydrase IV of new substituted 1,3,4-thiadiazole-, and 1,3,4-thiadiazoline-disulfonamides can be calculated in a short time, without any experiments. Future studies are necessary in order to assess the performances in computing the inhibition activity on carbonic anhydrase IV of new substituted 1,3,4-thiadiazole-disulfonamide or 1,3,4-thiadiazoline-disulfonamide.
CONCLUSIONS
The multi-varied MDF-SAR model with four descriptors is a solid and reliable one and indicates that the inhibition activity on carbonic anhydrase IV of substituted 1,3,4-thiadiazole- and 1,3,4-thiadiazoline-disulfonamides is like to be of geometry and topology nature, being in relation with compounds partial charges.
The MDF-SAR methodology opens a new pathway in computing and characterization of inhibitory activity on carbonic anhydrase IV of substituted 1,3,4-thiadiazole- and 1,3,4-thiadiazoline-disulfonamides.
REFERENCES
- 1.- Khadikar PV, Karmarkar S, Gour K, Agrawal VK, Singh S. QSAR study using distance-based topological indices. Oxidation Communications 2004;27(1):1-11.
2.- Allen DD, Geldenhuys WJ. Molecular modeling of blood-brain barrier nutrient transporters: in silico basis for evaluation of potential drug delivery to the central nervous system. Life Sci. 2006;78(10):1029-33.
3.- Verma RP, Mekapati SB, Kurup A, Hansch C. A QSAR review on melanoma toxicity. Bioorg Med Chem 2005;13(19):5508-26.
4.- Khadikar PV, Karmarkar S, Agrawal VK, Singh J, Shrivastava A, Lukovits I, Diudea MV. Szeged index - Applications for drug modeling. Letters in Drug Design and Discovery 2005;2 (8):606-624.
5.- Gutman I, Dobrynin AA. The Szeged index - A success story. Graph Theory Notes New York 1998;34:37-44.
6.- Nikolic S, Trinajstic N, Mihalic Z. The Wiener index: Development and applications. Croat Chem Acta 1995;68: 105-29.
7.- Yang F, Wang ZD, Huang YP. Modification of the Wiener index 4. J Comput Chem. 2004;25(6):881-7.
8.- Vukicevic´ D, Z?erovnik J. Altered Wiener indices. Acta Chimica Slovenica 2005;52(3):272-81.
9.- Khadikar PV, Karmarkar S, Agrawal VK. A Novel PI Index and its Applications to QSPR/QSAR Studies. J Chem Inf Comput Sci 2001; 41(4):934-949.
10.- Jaiswal M, Khadikar P. QSAR study on tadpole narcosis using PI index: a case of heterogenous set of compounds. Bioorg Med Chem 2004;12(7):1731-6.
11.- Thakur A, Thakur M, Khadikar PV, Supuran CT, Sudele P. QSAR study on benzenesulphonamide carbonic anhydrase inhibitors: topological approach using Balaban index. Bioorg Med Chem 2004;15;12(4):789-93.
12.- Nikolic S, Kovacevic´ G, Milicevic´ A, Trinajstic N. The Zagreb indices 30 years after. Croatica Chemica Acta 2003;76(2):113-24.
13.- Li XH, Jalbout AF, Solimannejad M. Definition and application of a novel valence molecular connectivity index. Journal of Molecular Structure: THEOCHEM 2003;663:81-5.
14.- Lu C, Guo W, Hu X, Wang Y, Yin C. A Lu index for QSAR/QSPR studies. Chem Phys Lett 2006;417(1-3):11-5.
15.- Lu C, Guo W, Wang Y, Yin C. Novel distance-based atom-type topological indices DAI for QSPR/QSAR studies of alcohols. J Mol Model (Online). 2006;1-8.
16.- Ren B. Application of novel atom-type AI topological indices to QSPR studies of alkanes. Comput Chem 2002;26(4):357-69.
17.- Baird TT Jr, Waheed A, Okuyama T, Sly WS, Fierke CA. Catalysis and inhibition of human carbonic anhydrase IV. Biochemistry 1997;36(9):2669-78.
18.- Clare BW, Supuran CT. A physically interpretable quantum-theoretic QSAR for some carbonic anhydrase inhibitors with diverse aromatic rings, obtained by a new QSAR procedure. Bioorg Med Chem. 2005;13(6):2197-211.
19.- Jaiswal M, Khadikar PV, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors: the first QSAR study on inhibition of tumor-associated isoenzyme IX with aromatic and heterocyclic sulfonamides. Bioorg Med Chem Lett. 2004;14(12):3283-90.
20.- Vullo D, Steffansen B, Brodin B, Supuran C, Scozzafava A, Nielsen C. Carbonic anhydrase inhibitors: Transepithelial transport of thioureido sulfonamide inhibitors of the cancer-associated isozyme IX is dependent on efflux transporters. Bioorg Med Chem 2006;14(7):2418-2427.
21.- Melagraki G, Afantitis A, Sarimveis H, Igglessi-Markopoulou O, Supuran CT. QSAR study on para-substituted aromatic sulfonamides as carbonic anhydrase II inhibitors using topological information indices. Bioorg Med Chem. 2006;14:1108-14.
22.- Supuran CT, Clare BW. Quantum theoretic QSAR of benzene derivatives: some enzyme inhibitors. J Enzyme Inhib Med Chem. 2004;19(3):237-48.
23.- Melagraki G, Afantitis A, Sarimveis H, Igglessi-Markopoulou O, Supuran CT. QSAR study on para-substituted aromatic sulfonamides as carbonic anhydrase II inhibitors using topological information indices. Bioorg Med Chem 2006;14(4):1108-14.
24.- Gupta SP, Kumaran S. A quantitative structure-activity relationship study on some aromatic/heterocyclic sulfonamides and their charged derivatives acting as carbonic anhydrase inhibitors. J Enzyme Inhib Med Chem. 2005;20(3):251-9.
25.- Pastorekova S, Parkkila S, Pastorek J, Supuran CT. Carbonic anhydrases: current state of the art, therapeutic applications and future prospects. J Enzyme Inhib Med Chem 2004;19(3):199-229.
26.- Masereel B, Rolin S, Abbate F, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors: anticonvulsant sulfonamides incorporating valproyl and other lipophilic moieties. J Med Chem 2002;45(2):312-20.
27.- Svichar N, Esquenazi S, Waheed A, Sly WS, Chesler M. Functional demonstration of surface carbonic anhydrase IV activity on rat astrocytes. Glia 2006;53(3):241-7.
28.- Supuran TC, Clare WB. Carbonic anhydrase inhibitors - Part 57: Qunatum chemical QSAR of a group of 1,3,4-thiadiazole- and 1,3,4-thiadiazoline disulfonamide with carbonic anhydrase inhibitory properties. Eur J Med Chem 1999;34:41-50.
29.- Jäntschi L. Molecular Descriptors Family on Structure Activity Relationships 1. The review of Methodology. Leonardo Electronic Journal of Practices and Technologies 2005;6:76-98.
30.- Jäntschi L. Delphi Client - Server Implementation of Multiple Linear Regression Findings: a QSAR/QSPR Application. Applied Medical Informatics 2004;15:48-55.
31.- Bolboaca S, Jäntschi L. Molecular Descriptors Family on Structure Activity Relationships 3. Antituberculotic Activity of some Polyhydroxyxanthones, Leonardo Journal of Sciences 2005;7:58-64.
32.- HyperChem , Molecular Modelling System [Internet page]; ©2003, Hypercube [about three screens]; [cited 2005 Nov]. Available from URL: http://hyper.com/products/
33.- Jäntschi L, Katona G, Diudea VM. Modeling Molecular Properties by Cluj Indices. Commun. Math. Comput. Chem. (MATCH) 2000;41:151-188.
34.- Diudea M, Gutman I, Jäntschi L. Molecular Topology, 2nd Edition, Nova Science, Huntington, New York, 2002.
35.- Leave-one-out Analysis. ©2005, Virtual Library of Free Software [cited 2006 March]. Available from URL:
http://vl.academicdirect.org/molecular_topology/mdf_findings/loo/.
36.- Steiger JH. Tests for comparing elements of a correlation matrix. Psychol Bull 1980;87:245-51.
37.- Training vs. Test Experiment. ©2005, Virtual Library of Free Software [cited 2006 March]. Available from URL:
http://vl.academicdirect.org/molecular_topology/qsar_qspr_s/.
38.- MDF SAR Predictor, © 2005, Virtual Library of Free Software [cited 2006 March]. Available from URL:
http://vl.academicdirect.org/molecular_topology/mdf_findings/sar.
ACKNOWLEDGEMENT: Research was in partly supported by UEFISCSU Romania through project ET36/2005
| Lorentz JÄNTSCHI
Technical University of Cluj-Napoca, 15 Constantin Daicoviciu Street, 400020 Cluj-Napoca, Romania http://lori.academicdirect.org |
Sorana Daniela BOLBOACA
|
Received, June 16, 2006. Received reviewed July 26, 2006
Publicado, 31 de Julio de 2006.
